Optimizing DNA Extraction and Selecting Suitable Regions for Biodiversity Assessment: A Study on Shorea leprosula
Abstract
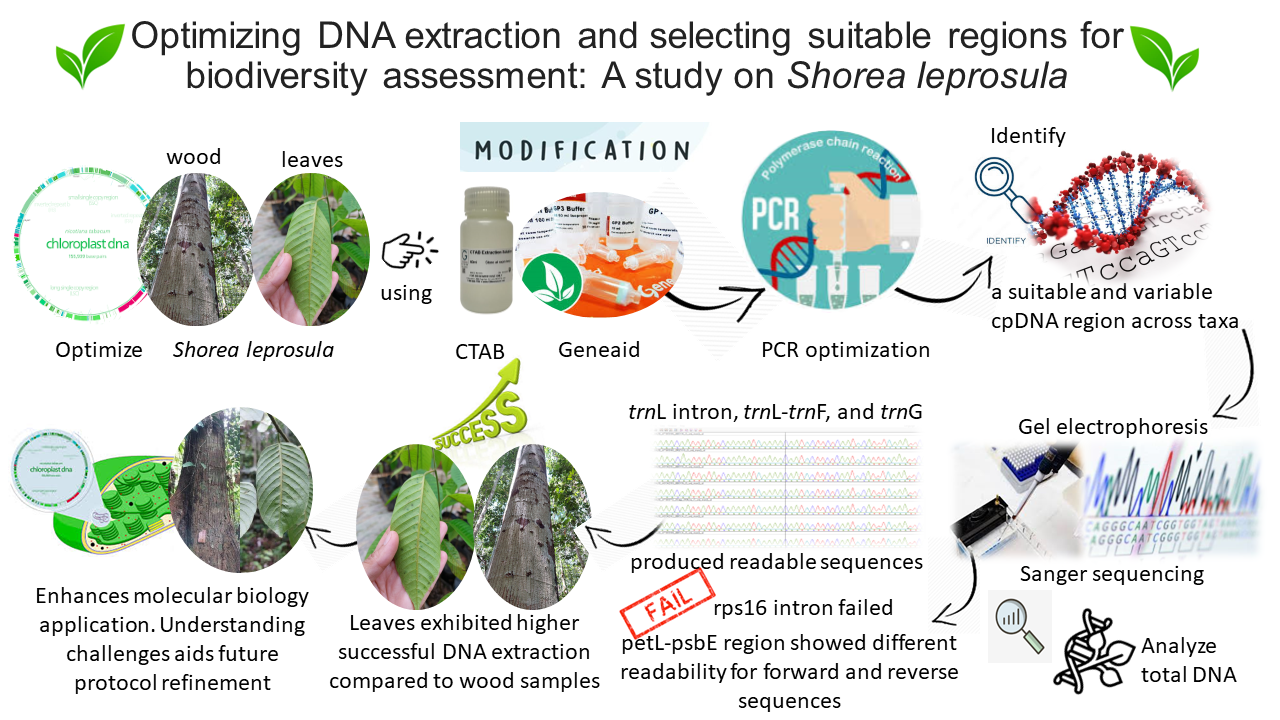
The extraction method plays a crucial role in obtaining high-quality DNA samples, which is indispensable for various molecular biology techniques and analyses, enabling a deeper comprehension of genetic information and biological processes. The objectives of the study were: a) to optimize the chloroplast DNA extraction protocol by comparing modified CTAB methods and GeneAid for both leaf and wood samples of Shorea leprosula, a major commercial timber species, and b) to identify a suitable cpDNA region that exhibits variability and universality across taxa. Total DNA was analyzed by gel electrophoresis followed by Sanger sequencing to determine the amplification success. The results revealed that trnL intron, trnL-trnF, and trnG yielded readable sequences of the expected length (maximum 586 bp, 480 bp, and 908 bp, respectively), while the rps 16 intron failed to assemble a contig. The petL-psbE region provided long readability for reverse sequences (769 bp) but not for the forward sequence (195 bp). Higher successful DNA extraction was achieved from the leaves compared to the woods. The lower sequencing quality may be attributed to suboptimal primer design, the structural features of the regions resulting from extensive repetitive sequences, and the suboptimal condition of the extraction method in eliminating wood chemical compounds.
References
Abasolo, M. A., Fernando, E. S., Borromeo, T. H., & Hautea, D. M. (2009). Cross-species amplification of Shorea microsatellite DNA markers in Parashorea malaanonan (Dipterocarpaceae). Philippine Journal of Science, 138(1), 23–28.
Abe, H., Watanabe, U., Yoshida, K., Kuroda, K., & Zhang, C. (2011). Changes in organelle and DNA quality, quantity, and distribution in the wood of Cryptomeria japonica over long-term storage. IAWA Journal, 32(2), 263–272. https://doi.org/10.1163/22941932-90000056
Ashton, P. S. (1982). Dipterocarpaceae. In Flora malesiana I, Spermatophyta (pp. 237–552). Rijksherbarium.
Bayer, R. J., Puttock, C. F., & Kelchner, S. A. (2000). Phylogeny of South African Gnaphalieae (Asteraceae) based on two noncoding chloroplast sequences. American Journal of Botany, 87(2), 259–272. https://doi.org/10.2307/2656914
Bayer, R. J., & Starr, J. R. (1998). Tribal phylogeny of the Asteraceae based on two non-coding chloroplast sequences, the trnL Intron and trnL/trnF Intergenic spacer. Annals of the Missouri Botanical Garden, 85, 242–256. https://doi.org/10.2307/2992008
Cannon, C. H., & Manos, P. S. (2003). Phylogeography of the Southeast Asian stone oaks (Lithocarpus). Journal of Biogeography, 30(2), 211–226. https://doi.org/10.1046/j.1365-2699.2003.00829.x
Cano, R. J. (1996). Analysing ancient DNA. Endeavour, 20(4), 162–167. https://doi.org/10.1016/S0160-9327(96)10031-4
Cao, C. P., Finkeldey, R., Siregar, I. Z., Siregar, U. J., & Gailing, O. (2006). Genetic diversity within and among populations of Shorea leprosula Miq. and Shorea parvifolia Dyer (Dipterocarpaceae) in Indonesia detected by AFLPs. Tree Genetics and Genomes, 2(4), 225–239. https://doi.org/10.1007/s11295-006-0046-0
Deguilloux, M. F., Pemonge, M. H., Bertel, L., Kremer, A., & Petit, R. J. (2003). Checking the geographical origin of oak wood: Molecular and statistical tools. Molecular Ecology, 12(6), 1629–1636. https://doi.org/10.1046/j.1365-294X.2003.01836.x
Deguilloux, M. F., Pemonge, M. H., & Petit, R. J. (2002). Novel perspectives in wood certification and forensics: Dry wood as a source of DNA. Proceedings of the Royal Society B: Biological Sciences, 269(1495), 1039–1046. https://doi.org/10.1098/rspb.2002.1982
Dellaporta, S. L., Wood, J., & Hicks, J. B. (1983). A plant DNA minipreparation: Version II. Plant Molecular Biology Reporter, 1(4), 19–21. https://doi.org/10.1007/BF02712670
Dhaliwal, A. (2020). DNA extraction and purification. Mater Methods, 3, 191. https://doi.org/10.13070/mm.en.3.191
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19(1), 11–15.
Eckert, K. A., & Kunkel, T. A. (1991). DNA polymerase fidelity and the polymerase chain reaction. Genome Research, 1(1), 17–24. https://doi.org/10.1101/gr.1.1.17
Filippis, L. De, & Magel, E. (1998). Differences in genomic DNA extracted from bark and from wood of different zones in Robinia trees using RAPD-PCR. Trees, 12(6), 377–384. https://doi.org/10.1007/PL00009723
Finkeldey, R., Leinemann, L., & Gailing, O. (2010). Molecular genetic tools to infer the origin of forest plants and wood. Applied Microbiology and Biotechnology, 85(5), 1251–1258. https://doi.org/10.1007/s00253-009-2328-6
Francis, F., Dumas, M. D., & Wisser, R. J. (2017). ThermoAlign: A genome-aware primer design tool for tiled amplicon resequencing. Scientific Reports, 7, 44437. https://doi.org/10.1038/srep44437
González, D., Vovides, A., & Lammers, T. G. (2002). Low intralineage divergence in Ceratozamia (Zamiaceae) detected with nuclear ribosomal DNA ITS and chloroplast DNA trnL-F non-coding region. Systematic Botany, 27(4), 654–661.
Gryson, N. (2010). Effect of food processing on plant DNA degradation and PCR-based GMO analysis: A review. Analytical and Bioanalytical Chemistry, 396(6), 2003–2022. https://doi.org/10.1007/s00216-009-3343-2
Haroen, W. K., & Dimyati, F. (2006). Sifat kayu tarik, teras, dan gubal Acacia mangium terhadap karakteristik pulp. Berita Selulosa, 41(1), 1–7.
Heckenhauer, J., Samuel, R., Ashton, P. S., Abu Salim, K., & Paun, O. (2018). Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreae (Dipterocarpaceae). Molecular Phylogenetics and Evolution, 127, 1–13. https://doi.org/10.1016/j.ympev.2018.05.010
Ibrahim, R. I. H. (2011). A modified CTAB protocol for DNA extraction from young flower petals of some medicinal plant species. Geneconserve, 10(40), 165–182.
Jhala Vibhuti, M., Mandaliya, V. B., & Thaker, V. S. (2015). Simple and efficient protocol for RNA and DNA extraction from rice (Oryza sativa L.) for downstream applications. International Research Journal of Biological Sciences, 4(2), 62–67.
Jobes, D. V, Hurley, D. L., & Thien, L. B. (1995). Plant DNA isolation: A method to efficiently remove polyphenolics, polysaccharides, and RNA. Taxon, 44, 379–386.
Kajita, T., Kamiya, K., Nakamura, K., Tachida, H., Wickneswari, R., Tsumura, Y., Yoshimaru, H., & Yamazaki, T. (1998). Molecular phylogeny of Dipetrocarpaceae in Southeast Asia based on nucleotide sequences of matK, trnL intron, and trnL-trnF intergenic spacer region in chloroplast DNA. Molecular Phylogenetics and Evolution, 10(2), 202–209. https://doi.org/10.1006/mpev.1998.0516
Kamiya, K., Gan, Y. Y., Lum, S. K. Y., Khoo, M. S., Chua, S. C., & Faizu, N. N. H. (2011). Morphological and molecular evidence of natural hybridization in Shorea (Dipterocarpaceae). Tree Genetics and Genomes, 7, 297–306. https://doi.org/10.1007/s11295-010-0332-8
Kamiya, K., Nanami, S., Kenzo, T., Yoneda, R., Diway, B., Chong, L., Azani, M. A., Majid, N. M., Lum, S. K. Y., Wong, K. M., & Harada, K. (2012). Demographic history of Shorea curtisii (Dipterocarpaceae) inferred from chloroplast DNA sequence variations. Biotropica, 44(5), 577–585. https://doi.org/10.1111/j.1744-7429.2011.00834.x
Karaca, M., İnce, A. G., Elmasulu, S. Y., Onus, A. N., & Turgut, K. (2005). Coisolation of genomic and organelle DNAs from 15 genera and 31 species of plants. Analytical Biochemistry, 343(2), 353–355. https://doi.org/10.1016/j.ab.2005.03.021
Kärkönen, A., & Koutaniemi, S. (2010). Lignin biosynthesis studies in plant tissue cultures. Journal of Integrative Plant Biology, 52(2), 176–185. https://doi.org/10.1111/j.1744-7909.2010.00913.x
Kawamura, F., Ramle, S. F. M., Sulaiman, O., Hashim, R., & Ohara, S. (2011). Antioxidant and antifungal activities of extracts from 15 selected hardwood species of Malaysian timber. European Journal of Wood and Wood Products, 69(2), 207–212. https://doi.org/10.1007/s00107-010-0413-2
Kessler, P. J. A., & Sidiyasa, K. (1994). Trees of the Balikpapan-Samarinda area, East Kalimantan, Indonesia: A manual to 280 selected species. Tropenbos Foundation.
Kim, C. S., Lee, C. H., Shin, J. S., Chung, Y. S., & Hyung, N. I. (1997). A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Research, 25(5), 1085–1086. https://doi.org/10.1093/nar/25.5.1085
Kim, D., Hofstaedter, C. E., Zhao, C., Mattei, L., Tanes, C., Clarke, E., Lauder, A., Sherrill-mix, S., Chehoud, C., Kelsen, J., Conrad, M., Collman, R. G., Baldassano, R., Bushman, F. D., & Bittinger, K. (2017). Optimizing methods and dodging pitfalls in microbiome research. Microbiome, 5, 52. https://doi.org/10.1186/s40168-017-0267-5
Kirchhoff, H. (2019). Chloroplast ultrastructure in plants. New Phytologist, 223(2), 565–574. https://doi.org/10.1111/nph.15730
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054
Lee, S. L., Wickneswari, R., Mahani, M. C., & Zakri, A. H. (2000a). Inheritance of allozyme in Shorea leprosula (Dipterocarpaceae). Journal of Tropical Forest Science, 12(1), 124–138.
Lee, S. L., Wickneswari, R., Mahani, M. C., & Zakri, A. H. (2000b). Genetic diversity of a tropical tree species, Shorea leprosula Miq. (Dipterocarpaceae), in Malaysia: Implications for conservation of genetic resources and tree improvement. Biotropica, 32(2), 213–224. https://doi.org/10.1111/j.1744-7429.2000.tb00464.x
Liepelt, S., Sperisen, C., Deguilloux, M. F., Petit, R. J., Kissling, R., Spencer, M., De Beaulieu, J. L., Taberlet, P., Gielly, L., & Ziegenhagen, B. (2006). Authenticated DNA from ancient wood remains. Annals of Botany, 98(5), 1107–1111. https://doi.org/10.1093/aob/mcl188
Liu, H., & Naismith, J. H. (2008). An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnology, 8, 91. https://doi.org/10.1186/1472-6750-8-91
Miranda Montero, J. J., Wright, E. M., & Khan, M. N. (2020). Illegal logging, fishing, and wildlife trade: The costs and how to combat it. World Bank Group. https://policycommons.net/artifacts/1282895/illegal-logging-fishing-and-wildlife-trade/
Mishra, G., Collings, D. A., & Altaner, C. M. (2018). Cell organelles and fluorescence of parenchyma cells in Eucalyptus bosistoana sapwood and heartwood investigated by microscopy. New Zealand Journal of Forestry Science, 48, 13. https://doi.org/10.1186/s40490-018-0118-6
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8(19), 4321–4326. https://doi.org/10.1093/nar/8.19.4321
Newman, M. F., Burgess, P. F., & Whitmore, T. C. (1996). Manual of dipterocarps for forester: Borneo Island light hardwoods, Anisptera, Parashorea, Shorea (red, white and yellow meranti). Royal Botanic Garden Edinburgh & CIFOR.
Ng, C. H., Ng, K. K. S., Lee, S. L., Zakaria, N. F., Lee, C. T., & Tnah, L. H. (2022). DNA databases of an important tropical timber tree species Shorea leprosula (Dipterocarpaceae) for forensic timber identification. Scientific Reports, 12, 9546. https://doi.org/10.1038/s41598-022-13697-x
Nuroniah, H. S., Gailing, O., & Finkeldey, R. (2010). Development of SCAR markers for species identification in the genus Shorea (Dipterocarpaceae). Silvae Genetica, 59(6), 249–257. https://doi.org/10.1515/sg-2010-0035
Okaura, T., & Harada, K. (2002). Phylogeographical structure revealed by chloroplast DNA variation in Japanese beech (Fagus crenata Blume). Heredity, 88(4), 322–329. https://doi.org/10.1038/sj.hdy.6800048
Pooma, R., & Newman, M. F. (2017). Shorea leprosula. https://doi.org/10.2305/ IUCN.UK.2017-3.RLTS.T33123A2833148.en
Porebski, S., Bailey, L. G., & Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, 15(1), 8–15. https://doi.org/10.1007/BF02772108
Rachmat, H. H., Kamiya, K., & Harada, K. (2012). Genetic diversity, population structure and conservation implication of the endemic Sumatran lowland dipterocarp tree species (Shorea javanica). International Journal of Biodiversity and Conservation, 4(14), 573–583. https://doi.org/10.5897/IJBC12.045
Rachmayanti, Y., Leinemann, L., Gailing, O., & Finkeldey, R. (2009). DNA from processed and unprocessed wood: factors influencing the isolation success. Forensic Science International. Genetics, 3(3), 185–192. https://doi.org/10.1016/j.fsigen.2009.01.002
Rahmadara, G., Hanifah, N. F., Rismayanti, R., Purwoko, D., Rochandi, A., & Tajuddin, T. (2022). Comparison of DNA isolation methods that derived from leaves of a potential anti-cancer rodent tuber (Typhonium flagelliforme) plant. International Journal of Agriculture System, 10(2), 93. https://doi.org/10.20956/ijas.v10i2.3966
Rana, R., Villarin, R., Gailling, O., Finkeldey, R., & Polle, A. (2013). Height growth, wood density and molecular markers to distinguish five tree species of Dipterocarpaceae grown at same site. Bangladesh Journal of Scientific and Industrial Research, 47(4), 407–414. https://doi.org/10.3329/bjsir.v47i4.14070
Risnasari, I., Nuryawan, A., & Siallagan, N. F. (2019). Characterization of particleboard from waste tea leaves (Camellia sinensis L) and meranti wood (Shorea sp.) using urea-formaldehyde adhesive and it’s formaldehyde emission. Proceedings of the International Conference on Natural Resources and Technology, 1, 261–264. https://doi.org/10.5220/0008552702610264
Rogers, S. O., & Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology, 5(2), 69–76. https://doi.org/10.1007/BF00020088
Rohland, N., & Hofreiter, M. (2007). Comparison and optimization of ancient DNA extraction. BioTechniques, 42(3), 343–352. https://doi.org/10.2144/000112383
Rudjiman, A. (2002). Identification manual of Shorea spp. ITTO Project-Faculty of Forestry Gadjah Mada University.
Sablok, G., Gahlot, P., Gupta, A. K., Pareek, K., & Sekhawat, N. S. (2009). Extraction of PCR-usable DNA from trees adapted to arid environment. Plant Omics Journal, 2(3), 103–109.
Sahu, S. K., Thangaraj, M., & Kathiresan, K. (2012). DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Molecular Biology, 2012, 205049. https://doi.org/10.5402/2012/205049
Särkinen, T., Staats, M., Richardson, J. E., Cowan, R. S., & Bakker, F. T. (2012). How to Open the Treasure Chest? Optimising DNA Extraction from Herbarium Specimens. PLoS ONE, 7(8), e43808. https://doi.org/10.1371/journal.pone.0043808
Shaw, J., Lickey, E. B., Beck, J. T., Farmer, S. B., Liu, W., Miller, J., Siripun, K. C., Winder, C. T., Schilling, E. E., & Small, R. L. (2005). The tortoise and the Hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany, 92(1), 142–166. https://doi.org/10.3732/ajb.92.1.142
Shaw, J., Lickey, E., Schilling, E., & Small, R. (2007). Comparison of whole chloroplast genome sequence to choose noncoding regions for phylogenetic studies in Angiosperms: The tortoise and the Hare III. American Journal of Botany, 94, 275–288. https://doi.org/10.3732/ajb.94.3.275
Stewart, C. N., & Via, E. L. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques, 14(5), 748–751.
Swetha, V. P., Parvathy, V. A., Sheeja, T. E., & Sasikumar, B. (2014). Isolation and amplification of genomic DNA from barks of Cinnamomum spp. Turkish Journal of Biology, 38(1), 151–155. https://doi.org/10.3906/biy-1308-5
Syafriana, V., Rachmatiah, T., & Utama, N. W. (2020). Aktivitas antibakteri ekstrak metanol kulit batang meranti sarang punai (Shorea parvifolia Dyer) terhadap Staphylococcus aureus dan Propionibacterium acnes. Jurnal Farmasi Udayana, 2020(Special Issue), 160170. https://doi.org/10.24843/jfu.2020.v09.i03.p04
Syamkumar, S., Jose, M., & Sasikumar, B. (2005). Isolation and PCR amplification of genomic DNA from dried capsules of cardamom (Elettaria cardamomum M.). Plant Molecular Biology Reporter, 23(4), 37–41. https://doi.org/10.1007/bf02788890
Symington, C. F. (1943). Foresters’ manual of dipterocarps. Malayan forest record no. 16. Kuala Lumpur: Penerbit Universiti Malaya.
Taberlet, P., Gielly, L., Pautou, G., & Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology, 17(5), 1105–1109. https://doi.org/10.1007/BF00037152
Tibbits, J. F. G., McManus, L. J., Spokevicius, A. V, & Bossinger, G. (2006). A rapid method for tissue collection and high-throughput isolation of genomic DNA from mature trees. Plant Molecular Biology Reporter, 24(1), 81–91. https://doi.org/10.1007/BF02914048
Treangen, T. J., & Salzberg, S. L. (2012). Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nature Reviews Genetics, 13(1), 36–46. https://doi.org/10.1038/nrg3117
Tsumura, Y., Kado, T., Yoshida, K., Abe, H., Ohtani, M., Taguchi, Y., Fukue, Y., Tani, N., Ueno, S., Yoshimura, K., Kamiya, K., Harada, K., Takeuchi, Y., Diway, B., Finkeldey, R., Na’iem, M., Indrioko, S., Ng, K. K. S., Muhammad, N., & Lee, S. L. (2011). Molecular database for classifying Shorea species (Dipterocarpaceae) and techniques for checking the legitimacy of timber and wood products. Journal of Plant Research, 124(1), 35–48. https://doi.org/10.1007/s10265-010-0348-z
Turaki, A. A., Ahmad, B., Magaji, U. F., Abdulrazak, U. K., Yusuf, B. A., & Hamza, A. B. (2017). Optimised cetyltrimethylammonium bromide (CTAB) DNA extraction method of plant leaf with high polysaccharide and polyphenolic compounds for downstream reliable molecular analyses. African Journal of Biotechnology, 16(24), 1354–1365. https://doi.org/10.5897/ajb2017.15942
Varma, A., Padh, H., & Shrivastava, N. (2007). Plant genomic DNA isolation: An art or a science. Biotechnology Journal, 2(3), 386–392. https://doi.org/10.1002/biot.200600195
Wagner, D. B., Furnier, G. R., Saghai-Maroof, M. A., Williams, S. M., Dancik, B. P., & Allard, R. W. (1987). Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proceedings of the National Academy of Sciences, 84(7), 2097–2100. https://doi.org/10.1073/pnas.84.7.2097
Wahyudi, A., Sari, N., Saridan, A., Cahyono, D. D. N., Rayan, Noor, M., Fernandes, A., Abdurachman, Apriani, H., Handayani, R., Hardjana, A. K., Susanty, F. H., Karmilasanti, Ngatiman, Fajri, M., Wiati, C. B., & Wahyuni, T. (2014). Shorea leprosula Miq and Shorea johorensis Foxw: Ekologi, silvikultur, budaya dan pengembangan. Balai BesarPenelitian Dipterokarpa.
Williams, C. E., & Ronald, P. C. (1994). PCR template-DNA isolated quickly from monocot and dicot leaves without tissue homogenization. Nucleic Acids Research, 22(10), 1917–1918. https://doi.org/10.1093/nar/22.10.1917
Xu, J., Aileni, M., Abbagani, S., & Zhang, P. (2010). A reliable and efficient method for total rna isolation from various members of spurge family (Euphorbiaceae). Phytochemical Analysis, 21(5), 395–398. https://doi.org/10.1002/pca.1205
Yulita, K. S., Bayer, R. J., & West, J. G. (2005). Molecular phylogenetic study of Hopea and Shorea (Dipterocarpaceae): Evidence from the trnL-trnF and internal transcribed spacer regions. Plant Species Biology, 20(3), 167—182. https://doi.org/10.1111/j.1442-1984.2005.00136.x
Zimmermann, J., Hajibabaei, M., Blackburn, D. C., Hanken, J., Cantin, E., Posfai, J., & Evans, T. C. (2008). DNA damage in preserved specimens and tissue samples: A molecular assessment. Frontiers in Zoology, 5, 18. https://doi.org/10.1186/1742-9994-5-18
Authors

This work is licensed under a Creative Commons Attribution 4.0 International License.
Jurnal Manajemen Hutan Tropika is an open access journal which means that all contents is freely available without charge to the user or his/her institution. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles in this journal without asking prior permission from the publisher or the author. This is in accordance with the Budapest Open Access Initiative (BOAI) definition of open access.