PORTECT: Porcine detection kit using CRISPR-CAS method as new innovation for rapid and accurate halal detection
DOI:
https://doi.org/10.29244/hass.2.1.18-20Keywords:
Biotechnology, CRISPR-Cas, Food verification, Halal certification, Halal industry, Porcine DNA detectionAbstract
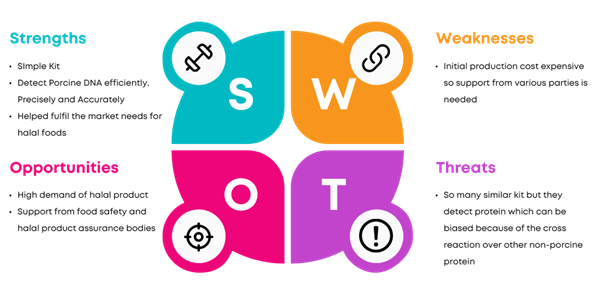
The PORTECT Porcine Detection Kit is an innovative solution leveraging CRISPR-Cas technology for rapid and accurate detection of porcine DNA in food products, addressing the growing demand for halal certification. This system uses a guide RNA (gRNA) designed specifically to recognize and target porcine DNA sequences, triggering Cas12 enzymes to cleave the DNA and a reporter compound for a visual color change. This process allows for precise, fast, and efficient detection of non-halal substances. Unlike traditional methods like PCR and ELISA, which require complex procedures, the PORTECT kit offers a more accessible and time-saving approach to halal verification, ensuring consumer trust and regulatory compliance. The technology is developed with bioinformatics tools and produces highly specific gRNA to target the porcine gene. The kit demonstrates significant potential in both the halal food industry and the general market for DNA detection tools, but future developments aim to lower production costs and expand detection capabilities beyond porcine DNA.
References
Ahmed I, Hossain MM, Fahim M, Rahman MS. Application of CRISPR-Cas systems in food authentication and halal food detection. Food Control. 2020;123:107763.
Alamillo JM, Lopez CM, Rivas FJ, Torralbo F, Bulut M, Alseekh S. Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein and hairy roots: a perfect match for gene functional analysis and crop improvement. Current Opinion in Biotechnology. 2023;79:102876. https://doi.org/10.1016/j.copbio.2022.102876
Briand L, Marvion G, Kriznik A, Heydel JM, Artur Y, Garrido C, Seigneuric R, Neiers F. A self-inducible heterologous protein expression system in Escherichia coli. Scientific Reports. 2016;6:33037. https://doi.org/10.1038/srep33037
Chen JS, Ma E, Harrington LB, Costa MD, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436-439. https://doi.org/10.1126/science.aar6245
Feng W, Zhang H, Le XC. Signal amplification by the trans-cleavage activity of CRISPR-Cas systems: Kinetics and performance. Analytical Chemistry. 2023. https://doi.org/10.1021/acs.analchem.2c04555
Gomez-Quintero OS, Morales-Moreno MD, Valdes-Galindo EG, Cardenas-Guerra RE. Optimizing heterologous production of CRISPR-AsCas12a protein in Escherichia coli. Research Square. 2024;1-18. https://doi.org/10.21203/rs.3.rs-4535821/v1
He W, Zhang X, Zou Y, Li J, Wang C, He Y, Jin Q, Ye J. Effective synthesis of high-integrity mRNA using in vitro transcription. Molecules. 2024;29(11). https://doi.org/10.3390/molecules29112461
Janudin AAS, Kurup CP, Chee YS, Mohd-Naim NF, Ahmed MU. Amplification-based CRISPR/Cas12a biosensor targeting the COX1 gene for specific detection of porcine DNA. ACS Omega. 2023;8(41):38212-38219. https://doi.org/10.1021/acsomega.3c04473
Karlsson M, Ekeroth J, Elwing H, Carlsson U. Reduction of irreversible protein adsorption on solid surfaces by protein engineering for increased stability. Journal of Biological Chemistry. 2005;280(27):25558-25564. https://doi.org/10.1074/jbc.M503665200
Khan MI, Haleem A. Understanding "halal" and "halal certification & accreditation system" - a brief review. Saudi Journal of Business and Management Studies. 2016;1(1):32-42.
Liu G, Zhang Y, Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Computational and Structural Biotechnology Journal. 2020;18:35-44. https://doi.org/10.1016/j.csbj.2019.11.006
Mohanraju P, Oost JD, Jinek M, Swarts DC. Heterologous expression and purification of CRISPR-Cas12a/Cpf1. Bio-protocol. 2018;8(9):1-23. https://doi.org/10.21769/BioProtoc.2842
Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120-1123. https://doi.org/10.1093/bioinformatics/btu743
Tyumentseva MA, Tyumentsev AI, Akimkin VG. Protocol for assessment of the efficiency of CRISPR/Cas RNP delivery to different types of target cells. PLoS One. 2021;16(11):e0260001. https://doi.org/10.1371/journal.pone.0259812
Wickramathilaka MP, Tao BY. Characterization of covalent crosslinking strategies for synthesizing DNA-based bioconjugates. Journal of Biological Engineering. 2019;13:63. https://doi.org/10.1186/s13036-019-0191-2
Wu Y, Dong Y, Shi Y, Yang H, Zhang J, Khan MR, Deng S, He G, He Q, Lv Y, Deng R. CRISPR-Cas12-based rapid authentication of halal food. Journal of Agricultural and Food Chemistry. 2021;69(35):10321-10328. https://doi.org/10.1021/acs.jafc.1c03078
Xiang Y, Ke W, Qin Y, Zhou B, Hu Y. PfAgo-based dual signal amplification biosensor for rapid and highly sensitive detection of alkaline phosphatase activity. Microchimica Acta. 2021;191:439. https://doi.org/10.1007/s00604-024-06516-9
Xu Y, Li Z. CRISPR-Cas systems: Overview, innovations, and applications in human disease research and gene therapy. Computational and Structural Biotechnology Journal. 2020;18:2401-2415. https://doi.org/10.1016/j.csbj.2020.08.031