Validation of porcine DNA analysis method for food products using selected primer and exogenous internal positive control in real-time PCR
Abstract
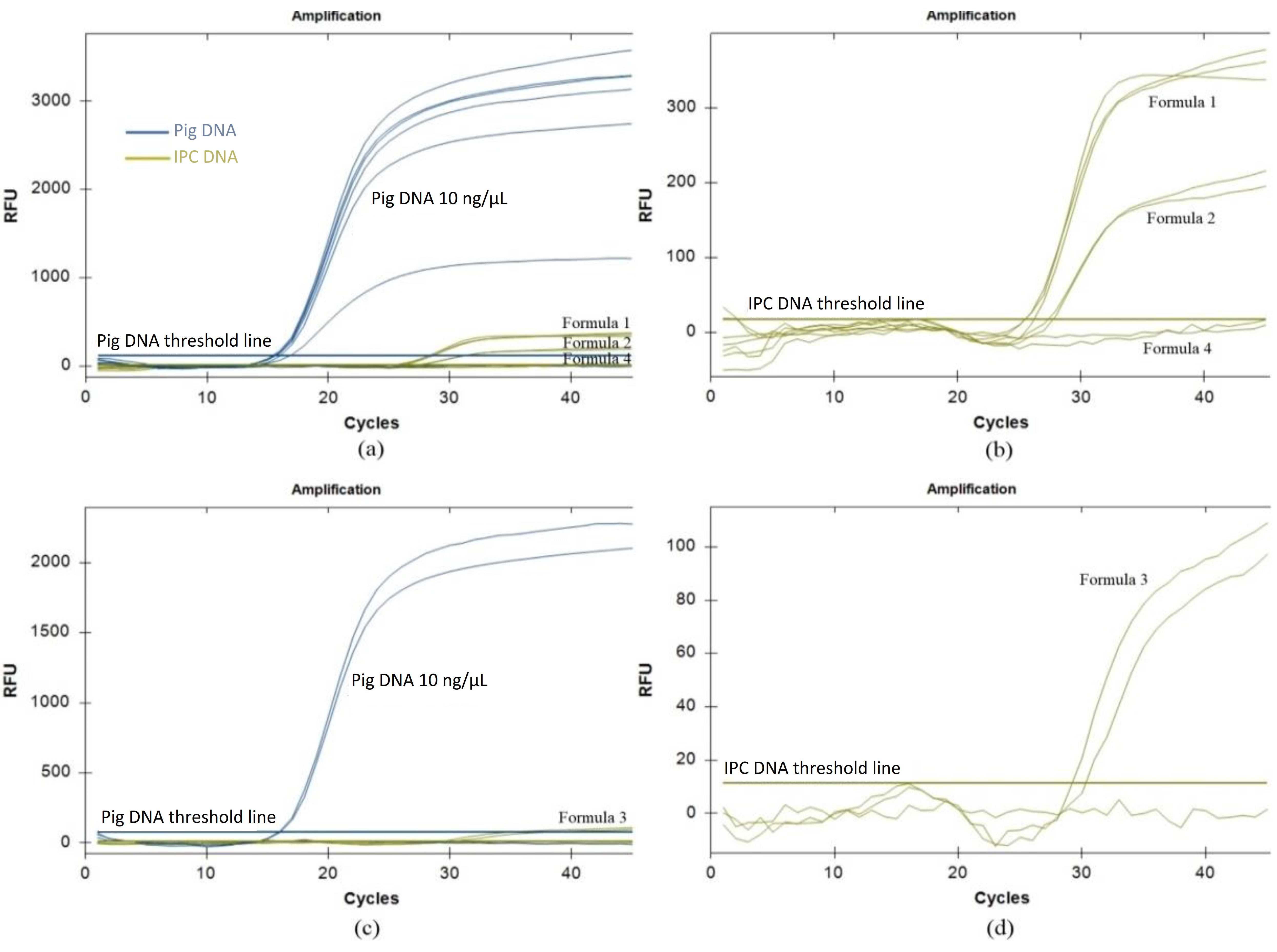
The method for porcine DNA analysis using real-time PCR is widely applied in the halal certification process and post-market monitoring. Therefore, this research aimed to validate porcine DNA analysis method using selected primer and exogenous internal positive control (IPC) as an alternative. The experiment was conducted in various stages, namely primer selection, sample extraction, efficiency testing, and method validation. The results of efficiency tests showed that using IPC at half concentration (Exo IPC Mix 5X and Exo IPC DNA 25X) provided reliable amplification with a Ct value of 27.57 ± 0.28 and RFU of 205.5 ± 14.85. The maximum DNA concentration that could be amplified without inhibition was 100 ng/µl. Validation tests showed specificity, sensitivity, linearity, PCR efficiency, and robustness. Among 23 positive and 23 negative samples, two positive samples (porcine collagen and collagen peptide) produced false negatives, while three negative samples had false positives after Ct 42.26. The method achieved a limit of detection (LOD) of 0.01 ng/l at Ct 33.29 ± 0.92, with linearity (r² = 0.996) and PCR efficiency (ϵ = 96.32). The results showed robustness to variations in master mix type, primer concentration, and annealing temperature, as well as resistance to inhibitors such as alginate, cellulose, EDTA, calcium ions, collagen peptide, and polysaccharides at 1 g/l. The performance of this method was also compared to the SNI ISO/TS 20224-3:2020 standard, showing potential as a viable alternative for porcine DNA testing. Moreover, further comparative research were recommended to fully establish the efficacy against national standards.
References
AppliedBiosystems. TaqPath™ ProAmp™ Master Mixes User Guide: Genotyping and Copy Number Variation PCR Workflows. Austin: Life Technologies Corporation; 2016.
Brezna B, Piknova L. Real-time PCR methods for identification of animal or plant species. In: Lazaro DR, editor. Real-time PCR in food science: current technology and applications. Burgos: Caister Academic Pr.; 2013. p. 253-271.
Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morisset D. Guidelines for validation of qualitative real-time PCR methods. Trends in Food Science & Technology. 2014;37:115-126. https://doi.org/10.1016/j.tifs.2014.03.008
Bustin S, Huggett J. qPCR primer design revisited. Biomolecular Detection and Quantification. 2017;14:19-28. https://doi.org/10.1016/j.bdq.2017.11.001
[BPOM]. Peraturan Kepala Badan Pengawas Obat dan Makanan tentang kategori pangan. Jakarta: No: 34; 2019.
Cahyaningsari D, Latif H, Sudarnika E. Identification of the addition of pork to beef-based foods using ELISA and qPCR. Animal Vaccines and Immunology. 2019;7(2):17-25. https://doi.org/10.29244/avi.7.2.17-25
Demirhan Y, Ulca P, Senyuva HZ. Detection of porcine DNA in gelatine and gelatine-containing processed food products-Halal/Kosher authentication. Meat Science. 2012;90:686-689. https://doi.org/10.1016/j.meatsci.2011.10.014
[FDA]. Guidelines for the validation of analytical methods for nucleic acid sequence-based analysis of food, feed, cosmetics and veterinary products. Washington: Food and Drug Administration; 2019.
Hedman J, Radstrom P. PCR detection of microbial pathogens: second edition, methods in molecular biology, vol. 943. Berlin: Springer Science+Business Media; 2013.
[ISO]. ISO 21571:2005 foodstuffs - methods of analysis for the detection of genetically modified organisms and derived products - nucleic acid extraction. Geneva: International Organization for Standardization; 2005.
Jansson L, Hedman J. Challenging the proposed causes of the PCR plateau phase. Biomolecular Detection and Quantification. 2019;17:1-7. https://doi.org/10.1016/j.bdq.2019.100082
Ke LD, Chen Z, Yung WK. A reliability test of standard-based quantitative PCR: exogenous vs endogenous standards. Molecular and Cellular Probes. 2000;14:127-135. https://doi.org/10.1006/mcpr.2000.0288
Kim S, Labbe RG, Ryu S. Inhibitory effects of collagen on the PCR for detection of Clostridium perfringens. Applied and Environmental Microbiology. 2000;66(3):1213-1215. https://doi.org/10.1128/AEM.66.3.1213-1215.2000
Lavanya DK, Sane A, Shivanna MB, Ganeshan S. Optimization of DNA isolation and PCR protocol for ISSR analysis of species of Spilanthes - a medicinal herb from Peninsular India. International Journal of Scientific Research. 2014;3(9):1088-1091.
Lazaro DR, Hernandez M. Introduction to the real-time polymerase chain reaction. In: Lazaro DR, editor. Real-time PCR in food science: current technology and applications. Burgos: Caister Academic Pr.; 2013. p. 3-19.
Lin J, Gu Y, Bian K. Bulk and surface chemical composition of wheat flour particles of different sizes. Journal of Chemistry. 2019:1-11. https://doi.org/10.1155/2019/5101684
Lorenz TC. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. Journal of Visualized Experiments. 2012;63:1-15. https://doi.org/10.3791/3998
[LPPOM MUI]. Work instructions for pig DNA extraction and analysis. Bogor: Institute for the Study of Food, Drugs and Cosmetics, Indonesian Ulema Council; 2019.
Martel F, Grundemann D, Schomig E. A simple method for elimination of false positive results in RT-PCR. Journal of Biochemistry and Molecular Biology. 2002;35(20):248-250. https://doi.org/10.5483/BMBRep.2002.35.2.248
McCord B, Pionzio A, Thompson R. Analysis of the effect of a variety of PCR inhibitors on the amplification of DNA using real time PCR, melt curves and STR analysis. Washington: Department of Justice; 2015.
[Menkes]. Keputusan Menteri Kesehatan Republik Indonesia tentang fortifikasi tepung terigu. Jakarta: No. 1452/MENKES/SK/X/2003; 2003.
Nakyinsige K, Man YBC, Sazili AQ. Halal authenticity issues in meat and meat products. Meat Science. 2012;91:207-214. https://doi.org/10.1016/j.meatsci.2012.02.015
Nurilmala M, Hizbullah HH, Karnia E, Kusumaningtyas E, Ochiai Y. Characterization and antioxidant activity of collagen, gelatin, and the derived peptides from yellowfin tuna (Thunnus albacares) skin. Marine Drugs. 2020;18(2):98. https://doi.org/10.3390/md18020098
Nurilmala M, Jacoeb AM, Dzaky RA. Characterization of fish skin gelatin yellowfin tuna. Jurnal Pengolahan Hasil Perikanan Indonesia. 2017;20(2):339-350. https://doi.org/10.17844/jphpi.v20i2.18049
Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. Journal of Applied Microbiology. 2012;113:1014-1016. https://doi.org/10.1111/j.1365-2672.2012.05384.x
Sefers S, Pei Z, Tang YW. False positives and false negatives encountered in diagnostic molecular microbiology. Medical Microbiology. 2005;16(2):59-67. https://doi.org/10.1097/01.revmedmi.0000166861.29699.2f
Tanabe S, Hase M, Yano T, Sato M, Fujimura T, Akiyama H. A real-time quantitative PCR detection method for pork, chicken, beef, mutton, and horseflesh in foods. Bioscience, Biotechnology, and Biochemistry. 2007;71(12):313-3135. https://doi.org/10.1271/bbb.70683
Tilaki KH. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian Journal of Internal Medicine. 2013;4(2):627-635.
Zilhadia, Izzah AN, Betha OS. Comparison of the SYBR Green and hydrolysis probe methods in the analysis of cow and pig gelatin DNA using real time PCR. Jurnal Sains dan Farmasi Klinis. 2017;4(2):16-23. https://doi.org/10.29208/jsfk.2017.4.1.194
Copyright (c) 2024 Heryani, Siti Nurjanah, Didah Nur Faridah

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.











