Detection of quinolone antibiotic resistance genes in Escherichia coli isolated from dairy cattle feces
DOI:
https://doi.org/10.29244/currbiomed.4.1.1Keywords:
antibiotic resistance genes, dairy cattle, DNA sequencing, Escherichia coli, quinolone resistanceAbstract
Background Livestock raised in densely populated areas can serve as reservoirs for bacteria such as Escherichia coli, which may harbor antibiotic resistance genes that threaten both animal and human health.
Objective This study aimed to identify and characterize quinolone resistance genes in E. coli isolated from dairy cattle feces.
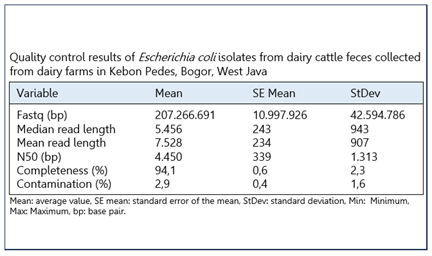
Methods Fifteen E. coli isolates were obtained from 15 dairy farms located in Kebon Pedes, Bogor, West Java. Genotypic detection of quinolone resistance genes was conducted using DNA sequencing on the MinION platform.
Results All E. coli isolates (100%) carried at least one quinolone resistance gene. Of these, ten isolates (67%) contained a single resistance gene, while five isolates (33%) possessed two genes. The qnrS1_1 gene was identified in all isolates and represented the predominant genotype, whereas the qnrVC4_1 gene was found in five isolates (33%), mostly co-occurring with qnrS1_1. Both genes are plasmid-mediated and categorized as plasmid-mediated quinolone resistance (PMQR) genes.
Conclusion The detection of qnrS1_1 and qnrVC4_1 genes in E. coli isolated from dairy cattle feces indicates that livestock manure may act as a reservoir for quinolone resistance genes, contributing to their persistence and potential spread within farm environments.
Downloads
Downloads
Published
Issue
Section
License
Copyright (c) 2026 Muhammad Ammar Raihan, Sara Elsharkawy, Agustin Indrawati, Hadri Latif

This work is licensed under a Creative Commons Attribution 4.0 International License.
This journal is published under a Creative Commons Attribution (CC BY 4.0) International License. This license enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creator.












